PCAT6
PCAT6: prostate cancer associated transcript 6, promotes cellular proliferation and colony formation of prostate cancer.
Contents
Annotated Information
Name
PCAT6: prostate cancer associated transcript 6 (HGNC:43714)
Aliases
ncRNA-a2; PCAN-R1: prostate cancer-associated noncoding RNA 1; KDM5BAS1: KDM5B antisense RNA 1 (head to head); onco-lncRNA-96 (HGNC:43714)
Characteristics
PCAT6 is a long non coding RNA gene located at chromosome 1q32.1, flanking the histone demethylase JARID1B/KDAM 5B. [1]
Function
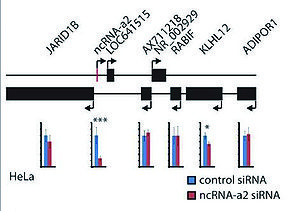
PCAT6 promotes cellular proliferation and colony formation of prostate cancer. Molecular analysis revealed that PCAT6 regulated the expression of two pivotal cancer-related proteins, c-Myc and p53, in lung cancer cells. [2]. Depletion of ncRNA-a2 (PCAT6) also results in significant reduction of KLHL12, a gene known for negative regulation of the Wnt-beta catenin pathway [1].
Expression
Detectable expression of ncRNA-a2 (PCAT6) was confirmed in HeLa cells. [1] PCAN-R1 or PCAT6 showed positive correlation between gene expression and somatic copy number alteration (SCNA). [3] Knockdown of PCAN-R1 resulted in substantial decrease in both cell growth and soft-agar colony formation in the androgen-dependent prostate cancer cell line LNCaP which was also later further confirmed in the androgen-independent prostate cancer cell line LNCaP-abl. The siRNA knockdown of PCAN-R1 had no effect on the expression of its neighboring protein-coding (PCG) gene KDM5B suggesting that the functional mechanism of PCAN-R1 is not directly through its neighboring protein coding genes. In normal tissues, PCAN-R1 and its neighboring PCG KDM5B showed the highest expression in testis [3]
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5′-CAGGAACCCCCTCCTTACTC-3 | 5′-CTAGGGATGTGTCCGAAGGA-3′[3] |
| RACE and northern blot analysis | 5′-GACCTGGGCAACCCCAGCCTG-3′ | 5′-GATCACTAATACGACTCACTATAGGGCGCGAGGAGCGCCTCATCACC-3′[3] |
| RNA interference | siR1-1 | 5′-GGTGTCTCCATCCTCATTC-3′[3] |
| siR1-2 | 5′-CTCCCAGACCTCACGTCAA-3′[3] |
Diseases
Labs working on this lncRNA
- Centre for Genomic Regulation (CRG), UPF, Barcelona, Spain [1]
- Center for Functional Cancer Epigeneitcs, Dana-Farber Cancer Institute, Boston, Massachusetts, USA [3]
- Department of Thoracic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China [2]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells[J]. Cell. 2010,143(1):46-58.
- ↑ 2.0 2.1 2.2 Wan L, Zhang L, Fan K, Cheng Z-X, Sun Q-C & Wang J-J. Knockdown of Long Noncoding RNA PCAT6 Inhibits Proliferation and Invasion in Lung Cancer Cells[J]. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2016, 24(3):161-170.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer[J]. Nature structural & molecular biology. 2013,20(7):908-13.
Sequence
>gi|375268763|ref|NR_046325.1| Homo sapiens prostate cancer associated transcript 6 (non-protein coding) (PCAT6), transcript variant 1, long non-coding RNA
000081 TAGGCGGGGT CAGGTTCGGG TTCCTAGGCC ATAGCGGAGC TGCAGCCCAG GCGGCCGGAG CGGAACCCAG CCCCGCTCCG 000160
000161 AGTGCCACGT CTCCAGGAAC CCCCTCCTTA CTCTTGGACA ACACTCCGCC CCCGCCCGGG CCTCCGTCCC CCAAACCGCC 000240
000241 CTCATTTGTG CAGCGCCAGA TCCTTCGGAC ACATCCCTAG GTGTCTCCAT CCTCATTCGG TCCATCCAAC TCCCAGACCT 000320
000321 CACGTCAACC GGCTGCACCC CACTTTCCAG CCTGCGCCCC AGATCTGCAG CCTTCGCCCC TAGATACACC CGCCTGGTGA 000400
000401 TGAGGCGCTC CTCGCGTTCC TTCCGGGCTC CAGGTGTCCG TGAGCCTCCC TTCCGCCCTG GCCTCCGGTC TCTGCCTTGC 000480
000481 TCGTGCTTCT ACCACCACCC TTCCCCTCCC AACCCGGTGG ATCCTCTCGT CTCCCCCAGT CTCCAGTGCA CCGGCTTTCC 000560
000561 CTCGTCCTCT GCGCAGTCCA TCTCAGCTCA TCTCTCCAAT TCAATGCCAT CATCTCTCCT CACCATCTCT CGGTGCCCTG 000640
000641 GAATGTTTGC TGTCAGATGT CCCCTGTGAA ACCCACAAAC GCTTGCGATT TGGCCTCCTT GTTTTATTTT GTGTAGTCCT 000720
000721 ACAACGTCTT GTTACTACCC CCTATTACAA CACTTATAAC TCAG