Help:Contents
Contents
FAQ
Introduction
- What is LncRNAWiki?
- LncRNAWiki is a wiki-based, publicly editable and open-content platform for community curation of human long non-coding RNAs (lncRNAs), viz., a community-curated lncRNA knowledgebase. Unlike conventional biological databases based on expert curation, lncRNAWiki harnesses collective intelligence to collect, edit and annotate information about lncRNAs, quantifies users' contributions in each annotated lncRNA and provides explicit authorship to encourage more participation from the whole scientific community.
You can perform different types of contributions to make LncRNAWiki the online encyclopedia for lncRNA.
- If you are a researcher, please share your knowledge and curate genes in your area of expertise.
- If you are a teacher/investigator, community curation of lncRNA genes in LncRNAWiki can be incorporated as student assignments, where contribution can be quantified as a score.
- If you are a student, you can work as a volunteer, e.g., data collection, content formatting.
- If you are a journal publisher, please consider community curation as a compulsory post-publication when any lncRNA-related paper is accepted by the journal.
- At the very least, please spread this news to any one who might be of interest.
- You can perform different types of contributions to make LncRNAWiki the online encyclopedia for lncRNA.
- How do I cite lncRNAWiki?
- The citation for lncRNAWiki is:
- LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs, submitted and under review.
- Other related publications:
- On the classification of long non-coding RNAs, RNA Biology, 2013, 10(6),925-933.
- AuthorReward: increasing community curation in biological knowledge wikis through automated authorship quantification, Bioinformatics, 2013, 29(14):1837-1839.
- RiceWiki: a wiki-based database for community curation of rice genes, Nucleic Acids Research, 2014,42(D1),D1222-D1228.
- Community intelligence in knowledge curation: an application to managing scientific nomenclature, PLoS ONE, 2013, 8(2):e56961.
- The citation for lncRNAWiki is:
LncRNAWiki Accounts
- Is it allowed for any user to provide edits in lncRNAWiki?
- LncRNAWiki allows any user to view and search but only registered users can add and edit content.
- Why do I open my identity and become a registered user?
- Open identity provided by registration not only improves content reliability, increases users’ collaborations and communications, but is also supportive to reward community-curated efforts by giving explicit authorship. It is of crucial significance for lncRNAWiki that would like to give credits to all contributors in reward for community-provided contents.
- How do I acquire a LncRNAWiki account?
- Please email the LncRNAWiki Team at lncwiki@big.ac.cn to tell us your preferred login name, real name, research interests, etc., and we will set up an account for you.
- How do I update my account information or change password?
- To update your account information, please log on first and then you may find a link named "My preferences" at the top right.
- Why I can't log into lncRNAWiki?
- Make sure that the Caps Lock key is not depressed. Passwords are case sensitive.
- Make sure your browser is set to accept cookies.
- Contact us at lncwiki@big.ac.cn.
Editing tips
- Formatting: http://www.mediawiki.org/wiki/Help:Formatting
- Images: http://www.mediawiki.org/wiki/Help:Images
- Links: http://www.mediawiki.org/wiki/Help:Links
- Tables: http://www.mediawiki.org/wiki/Help:Tables
- Lists: http://www.mediawiki.org/wiki/Help:Lists
Database content
Annotated Information
Annotated Information is organized as free text and of great helpfulness for users who share their knowledge and contribute edits without training in the curation or wiki techniques. It can also fall into several sub-sections, such as Function, Evolution, Expression, making it convenient to direct users to the sub-section(s) of interest. Although these sub-sections are preset, new sub-section can be easily added and irrelevant sub-section(s) can be deleted.
- The nomenclature of locus-specific transcripts
Example: N07QT0022001-SABC-LHPXX02 indicates the first non-coding transcript on the q-arm of chromosome 7 positioned on the 22th bin toward telomere, which resides in ABC gene locus in a sense direction, is a long transcript and highly expressed, and has an alternative promoter.
A locus-specific nomenclature is proposed as accession number for transcripts. It contains four consecutive segments joined by hyphens.
- Segment 1 contains 12 positions that display genomic information. The first three positions define the nature of a transcript and its residing chromosome. Capital letter N and C are dedicated to non-coding and coding transcripts, respectively. Chromosome numbers are indicated with two positions and 0 can be added to take the vacant position when the chromosome number is less than two digits (such as 01 and 07) or when X and Y are encountered (0X and 0Y are used). Transcription direction is, defined by the fourth and fifth positions, indicated with Q for the q-arms, P for the p-arms (when the transcript is on the centromere or telomere as well as subtelomeric and centromeric regions, the position is labeled as 0 rather than the definite Q or P), T for toward telomere, and C for toward centromere. The next seven digits are devoted to position the transcript; the first four define the bin number where the gene locus resides and the rest three define the number of transcripts within the bin based on first-come-first-name rule. The bins of a given chromosome are named from centromere to telomere in a nominal size of 100 Kb.To obtain the centromere location of each chromosome, you can refer to Media:Hg19cytoBand.txt.
- Segment 2 has four positions that define a gene locus. At the first position, S is used to indicate a non-coding transcript overlaps with a protein-coding gene on the same strand or in the same direction. A is used to indicate a non-coding transcript overlapping with the protein-coding gene on the antisense strand or in the opposite direction. For a protein-coding locus, the gene name is directly adapted. For a non-coding transcript, either the name of a nearby gene or a named non-coding RNA locus is abbreviated. When non-coding RNA has both S and A relationships with different protein-coding genes, S relationship is considered with priority.
- The last segment is used to define the characteristics of all transcripts in a locus. Seven positions are included: (1) transcript size is defined as large (L, >500 bp), mediate (M, from 100 bp to 500 bp), and small (S, from 20 bp to 100 bp). (2) Expression level is defined in three categories that include highly (H, RPKM or similar units>100), moderate (M, from 10 to 100), and low (L, <10). For lncRNAs that are differentially expressed, the highest expression level is considered. (3) Alternative promoter (P), alternative exon (E), alternative poly (A) are also indicated for all transcripts in the locus. When one or two of the three alternative forms are absent, X can be filled in to take the position(s). (4) Two digits are dedicated to accommodate the number of transcriptional variants.
Basic Information
Basic information includs 10 sub-sections, ‘Transcript ID’, ‘Source’, ‘Same with’, ‘Classification’, ‘Length’, ‘Genomic location’, ‘Exon number’, ‘Exons’, ‘Genome context’, and ‘Sequence’.
- Transcript ID: The original lncRNA ID in each database.
- Source: The database and version that this lncRNA is from.
- Same with: LncRNAs that have the same sequence and also the same genomic location in other databases.
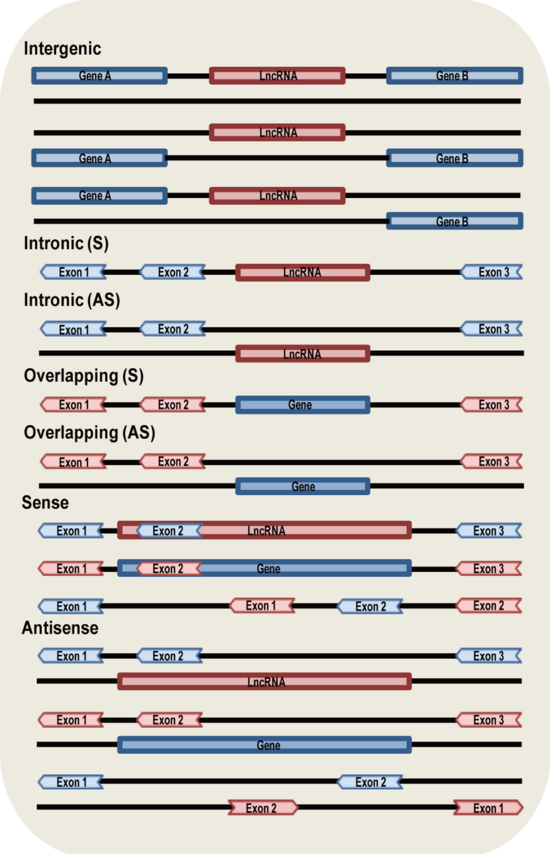
- Classification: Classification based on genomic location and context. We obtained genome location information from GENCODE, NONCODE and LNCipedia. Based on the categories of Derrien et al. (Derrien, T. et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res), we classified lncRNAs into seven groups (Intergenic, Intronic (S),Intronic (AS),Overlapping (S),Overlapping (AS), Sense and Antisense) based on their genomic location in respect to protein-coding genes. The difference between our classification and Derrien’s is that we classified lncRNAs that intersect protein-coding genes into Sense or Antisense by considering the whole transcript sequence instead of exonic region only.Most of the lncRNAs belong to only one category, a small number (1,264) belong to more than one category.
- Intergenic: lncRNAs are transcribed from intergenic regions.
- Intronic (S): lncRNAs are transcribed entirely from introns of protein-coding genes.
- Intronic (AS): lncRNAs are transcribed from antisense strand of protein-coding genes and the entire sequences are covered by introns of protein-coding genes.
- Overlapping (S): lncRNAs that contain coding genes within an intron on the sense strand.
- Overlapping (AS): lncRNAs that contain coding genes within an intron on the antisense strand.
- Sense: lncRNAs are transcribed from the sense strand of protein-coding genes and the entire sequence of lncRNAs are covered by protein-coding genes (Intronic lncRNAs are not included), or the entire sequence of protein-coding genes are covered by lncRNAs (Overlapping lncRNAs are not included ), or both lncRNAs and protein-coding genes intersect each other partially.
- Antisense: lncRNAs are transcribed from the antisense strand of protein-coding genes and the entire sequence of lncRNAs are covered by protein-coding genes (Intronic lncRNAs are not included), or the entire sequence of protein-coding genes are covered by lncRNAs (Overlapping lncRNAs are not included ), or both lncRNAs and protein-coding genes intersect each other partially.
- Length, Genomic location, Exon number and Exons: The length, genomic location, exon number of lncRNA, and genomic location of each exon. These information is obtained from GENCODE, NONCODE and LNCipedia annotation.
- Genome context: We integrated JBrowse (version 1.11.4) (http://jbrowse.org/) into LncRNAWiki to facilitate visualization of the genomic context and transcript structure of each lncRNA.
- Sequence: The transcript sequence of this lncRNA.
Predicted Small Protein
Predicted small protein (proteins of 100 amino acids or less in the absence of processing) includs 13 sub-sections, 'Name', 'Length(aa)', 'Molecular weight', 'Aromaticity', 'Instability index', 'Isoelectric point', 'Runs', 'Runs residual', 'Runs probability', 'Amino acid sequence', 'Secondary structure', 'PRMN', and 'PiMo'.
- Name: The name of predicted small proteins.
- Length (aa), Molecular weight, Aromaticity, Instability index, Isoelectric point: The length (aa), molecular weight, aromaticity, instability index, isoelectric point of predicted small proteins.
- Runs: The runs of secondary structure of predicted small proteins.
- Runs residual: The residual between runs/length of the predicted small protein and the average runs/length of mRNAs which have the same length with the predicted small protein.
- Runs probability: The average probability of the predicted small protein runs.
- Secondary structure: The amino acid sequence and its secondary structure of the predicted small protein.
- PRMN: Refined predicted transmembrane helix.
- PiMo: Predicted transmembrane region.