Difference between revisions of "ENST00000553157.1"
Chunlei Yu (talk | contribs) (→References) |
Chunlei Yu (talk | contribs) (→Labs working on this lncRNA) |
||
| Line 65: | Line 65: | ||
==Labs working on this lncRNA== | ==Labs working on this lncRNA== | ||
| − | + | * Centre for Genomic Regulation (CRG), UPF, Barcelona, Spain <ref name="ref1" /> | |
| + | |||
| + | |||
| + | * Center for Functional Cancer Epigeneitcs, Dana-Farber Cancer Institute, Boston, Massachusetts, USA <ref name="ref2" /> | ||
==References== | ==References== | ||
Revision as of 09:18, 24 June 2016
PCAT6
Contents
Annotated Information
Name
PCAT6: prostate cancer associated transcript 6 (non-protein coding)
ncRNA-a2, PCAN-R1, KDM5BAS1, onco-lncRNA-96
Characteristics
Function
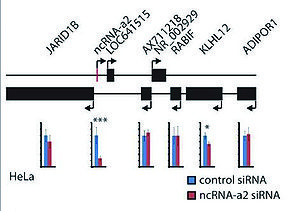
With the depletion of ncRNA-a2, the KLHL12, a gene known for its negative regulation of the Wnt-beta catenin pathway, displayed a significant reduction [1].
Expression
PCAN-R1 showed positive correlation between gene expression and SCNA. [2]
PCAN-R1, while one isoform PCAN-R1-A was almost identical to the Ensembl annotated transcript ENST00000425295, the other isoform PCAN-R1-B was a spliced variant of PCAN-R1-A with an intron retention. [2]
PCAN-R1 and its neighboring PCG KDM5B showed the highest expression in normal testis. [2]
| Primer | Forward primer | Reverse primer |
|---|---|---|
| RT-PCR | 5′-CAGGAACCCCCTCCTTACTC-3 | 5′-CTAGGGATGTGTCCGAAGGA-3′[2] |
| RACE and northern blot analysis | 5′-GACCTGGGCAACCCCAGCCTG-3′ | 5′-GATCACTAATACGACTCACTATAGGGCGCGAGGAGCGCCTCATCACC-3′[2] |
| RNA interference | siR1-1 | 5′-GGTGTCTCCATCCTCATTC-3′[2] |
| siR1-2 | 5′-CTCCCAGACCTCACGTCAA-3′[2] |
Conservation
Please input conservation information here.
Misc
Please input misc information here.
Transcriptomic Nomeclature
Please input transcriptomic nomeclature information here.
Regulation
Please input regulation information here.
Allelic Information and Variation
Please input allelic information and variation information here.
Evolution
Please input evolution information here.
You can also add sub-section(s) at will.
Labs working on this lncRNA
- Centre for Genomic Regulation (CRG), UPF, Barcelona, Spain [1]
- Center for Functional Cancer Epigeneitcs, Dana-Farber Cancer Institute, Boston, Massachusetts, USA [2]
References
- ↑ 1.0 1.1 1.2 Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells[J]. Cell. 2010,143(1):46-58.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer[J]. Nature structural & molecular biology. 2013,20(7):908-13.
Basic Information
| Transcript ID |
ENST00000553157.1 |
| Source |
Gencode19 |
| Same with |
lnc-RP11-480I12.4.1-1:2 |
| Classification |
{{{classification}}} |
| Length |
551 nt |
| Genomic location |
chr1+:202780082..202781284 |
| Exon number |
2 |
| Exons |
202780082..202780498,202781151..202781284 |
| Genome context |
|
| Sequence |
000001 AGAGGCCGGA CCTGGGCAAC CCCAGCCTGG AGGTGCCGGG GCCGGAGCTC CCAGAGGGCT GGGTGCGAGG CCTAGGCGGG 000080
000081 GTCAGGTTCG GGTTCCTAGG CCATAGCGGA GCTGCAGCCC AGGCGGCCGG AGCGGAACCC AGCCCCGCTC CGAGTGCCAC 000160 000161 GTCTCCAGGA ACCCCCTCCT TACTCTTGGA CAACACTCCG CCCCCGCCCG GGCCTCCGTC CCCCAAACCG CCCTCATTTG 000240 000241 TGCAGCGCCA GATCCTTCGG ACACATCCCT AGGTGTCTCC ATCCTCATTC GGTCCATCCA ACTCCCAGAC CTCACGTCAA 000320 000321 CCGGCTGCAC CCCACTTTCC AGCCTGCGCC CCAGATCTGC AGCCTTCGCC CCTAGATACA CCCGCCTGGT GATGAGGCGC 000400 000401 TCCTCGCGTT CCTTCCGGCA CGTGGCACAC GCTCCACAAA TATTTGTTGA AAGGGAGGGT AAATGGCCTT GAGTTTCTGT 000480 000481 GGCTACCATT GGCTGAGGAT GGAGCTTTGG GCTGGACAAA GAAGAATCTG GTTGACCACA TGAAGCAGGA A |