Difference between revisions of "ENST00000418282.1"
Chunlei Yu (talk | contribs) (→Characteristics) |
Chunlei Yu (talk | contribs) (→Transcriptomic Nomenclature) |
||
| Line 2: | Line 2: | ||
==Annotated Information== | ==Annotated Information== | ||
| − | === | + | ===Name=== |
''BPESC1'': blepharophimosis, epicanthus inversus and ptosis, candidate 1A (non-protein coding)(HGNC nomenclature) | ''BPESC1'': blepharophimosis, epicanthus inversus and ptosis, candidate 1A (non-protein coding)(HGNC nomenclature) | ||
Revision as of 06:13, 25 June 2016
BPESC1 ( BPES candidate 1), which contains three exons and spans a genomic fragment of 17.5 kb, is 3518 bp in size and contains an open reading frame of 351 bp.
Contents
Annotated Information
Name
BPESC1: blepharophimosis, epicanthus inversus and ptosis, candidate 1A (non-protein coding)(HGNC nomenclature)
NCRNA00187, "non-protein coding RNA 187"[1]
Characteristics
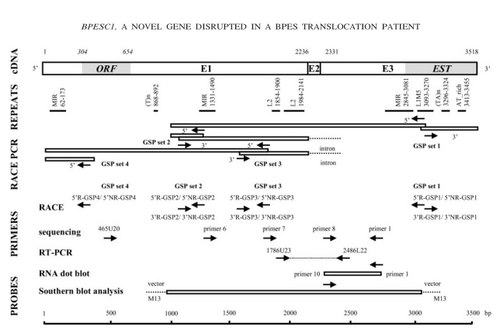
BPESC1 ( BPES candidate 1), a novel candidate gene that is disrupted by the translocation on chromosome 3. BPESC1 is 3518 bp in size and contains an open reading frame of 351 bp. BPESC1 contains three exons and spans a genomic fragment of 17.5 kb.[1]
Function
BPESC1 may play a direct or an indirect role in the clinical features of the t(3;4) BPES patient.[1].
Expression
BPESC1 is expressed neither in the adult ovary nor in the early fetal tissue might suggest its role in this specific developmental disorder.[1]
| Gene-specific primers(GSP) | PCR | Forward | Reverse |
|---|---|---|---|
| Primer set 1 | RACE | 5'R-GSP1(5'-AAC-TCA-CAC-CTA-CTC-CTG-TTT-CG-3') | 3'R-GSP1(5'-ATA-ATA-GGG-AGG-TTG-GGG-AAG-TG-3')[1] |
| Nested RACE | 5'NR-GSP1(5'-CCT-ACT-CCT-GTT-TCG-TCT-GTA-CC-3') | 3'NR-GSP1(5'-TTG-GGG-TAC-AGA-CGA-AAC-AGG-AG-3')[1] | |
| Primer set 2 | RACE | 5'R-GSP2(5'-TGT-GGC-CGG-AGG-AAC-TGA-GTG-AC-3') | 3'R-GSP2(5'-CCC-GAG-GCC-TTG-ACT-GGT-GAG-AT-3')[1] |
| Nested RACE | 5'NR-GSP2(5'-AGA-CTC-TGT-GTG-GGG-ACC-TTC-AT-3') | 3'NR-GSP2(5'-AGG-CCT-TGA-CTG-GTG-AGA-TTA-GC-3') | |
| Primer set 3 | RACE | 5'R-GSP3(5'-AAT-AAA-ATG-CCC-ACA-GGA-GAG-TT-3') | 3'R-GSP3(5'-GAG-CCC-CCA-GAT-TCT-CAC-CTT-T-3')[1] |
| Nested RACE | 5'NR-GSP3(5'-GGG-GAC-ATG-GAA-GAG-GAA-GAC-AA-3') | 3'NR-GSP3(5'-TTT-CTT-CTG-CCA-GGG-TTC-TTC-3')[1] | |
| Primer set 4 | RACE | 5'R-GSP4(5'-GAG-TAG-CCC-CAA-GGA-TGT-GTC-3')[1] | |
| Nested RACE | 5'NR-GSP4(5'-CTC-CCT-CAC-TCC-CAA-GAT-TTG-TG-3')[1] |
Labs working on this lncRNA
- Department of Medical Genetics, Ghent University Hospital, B-9000, Ghent, Belgium [1].
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 De Baere E, Fukushima Y, Small K, Udar N, Van Camp G, Verhoeven K, et al. Identification of BPESC1, a novel gene disrupted by a balanced chromosomal translocation, t(3;4)(q23;p15.2), in a patient with BPES[J]. Genomics. 2000,68(3):296-304.
Basic Information
| Transcript ID |
ENST00000418282.1 |
| Source |
Gencode19 |
| Same with |
lnc-C3orf72-1:1,BPESC1 |
| Classification |
intergenic |
| Length |
3518 nt |
| Genomic location |
chr3+:138823027..138844009 |
| Exon number |
3 |
| Exons |
138823027..138825263,138828054..138828147,138842823..138844009 |
| Genome context |
|
| Sequence |
000001 AAAGGAAGAA GAGGAACACG TTGCTGTGTG CTCGTGGCAC GTGGCCTGTT GGGTCCTTTC GCAGTCCTTT GAAGCAGTTG 000080
000081 CTTTTACTGT CCCCTGTTTT ATAGAAGAGG AAACAGCCAA GGTGACTTAA GGGCAGAGCG CCAGCAGGTA TGGAGCCTGA 000160 000161 ATTTAAATGC AGGTCTGCAT ACTGCAAGCA CCTGGGTTCT CTCCACCCCT CCTCTGGACT CCCATTTGTT CTCTGTCAGT 000240 000241 GACTGAGAAA GGGTGGGTGG GACTAGGAAC TCAGCTTCCG GTCCATGACC ACATCCATGG AATATGGGAG GAAGAGATCA 000320 000321 AATCACAAAT CTTGGGAGTG AGGGAGAGAC ACATCCTTGG GGCTACTCAA GCCCCCCATA TCCTTTGCAT ACCTTCTTTA 000400 000401 TCCCTTTTCT ACCCAGTGGA TTTGGAGGGA GTGGGCTTGG GATACCATCA GACAACGAGA AACACGATTT GCAGGACTGT 000480 000481 GTGGAGGTAT CCAGGCCTGA GGGCCCTGCT CCAGAGCTCC CCTCCTCACT CTGTGGCTGG AACAAAATCT CTTCCTTGTG 000560 000561 TGGCCTTGGC TTCCCTAGCA GGGATCCCAA AACATGGGAT CTGGCCATGC TGCTCAGTCC TTGGGTTGAT TTCTTTGAGC 000640 000641 TCCAGCTTCT CTGAGCATCC CCTCTGCTGT CCCCACATCC TGATAGCCCC TCTGGGAGCT GGGGCCTGGC CTTTCTGCCC 000720 000721 ATTCTCAACA AACACTCTTG TCTGGAGACA GGCAGGACAA TGGAGAGCTA CCTTGTCCTG CTGCTGCCCC TGCCACCACA 000800 000801 GCTGCTTATG TGTGACCGAT GGGGTTGAGA GGGGCAAGAG TTCTGTGGAG AGAGTGGAGG GAAAGAGTTT TTTTTTTTTT 000880 000881 TTTTTTTTTT TTTGCTATGC ACAGTTCCTC CCAGCAACCT TCCCTCTCTG ACCAATGGGG CCTCTAGGCT CTGCCCTTCA 000960 000961 CGTTGGGGTC TTTCCCCACC TCCGGCCCTT TGGTTTTGAT ATGGCTAGTG ACCAGCACTG ACCACCATCC CAAGCTGGGA 001040 001041 AGAGCAATTG GTGAAGAGTG TGTGTACTAT TTACTTTCTT CACCCTTGCC CCTTTATTCT CAAGGTCTTT TGATTGTGAA 001120 001121 GCAAGGCCCG AGGCCTTGAC TGGTGAGATT AGCCCGGGAG GTGGTGGCTC CCTCCTGCTC TCTGTGGTGA AAGCCAAGCA 001200 001201 CACTTTCCTA AAACCAGCGT CGGAAGCTGT GCTGTAGCCT GTAATTACTG ACATGAAGGT CCCCACACAG AGTCTGGATG 001280 001281 GAGGCTGCCC TGGAAGCAGC CCCGGGCTGG GAAAGGGCTG AGTGTCAACA GGCCTGTGTT TGAATCCAAG TCACCTGTGT 001360 001361 TCTCTGACAG GTCACTCAGT TCCTCCGGCC ACAGCTGCCC CTCTGTGAGA TGACATAACA CTGCTTACCT CTTGGTGTGG 001440 001441 CTATGAGGAT TGATTGGGCT GATGTGTAGA AAATATGTAA TGCAGGGCTT GTCCTGGAGG TGATGTTAGG TAATGGTGAG 001520 001521 GTGGATCTTC CCTGTGTGGT CCCTCCCTTC GTGTGCCCCT GCCCAGCCTC TTGGCACCTC CTGTTTACTC CCCGAGCCCC 001600 001601 CAGATTCTCA CCTTTGCCCA GAGCCACATT CGGGCTTGTC TTCCTCTTCC ATGTCCCCGC ATGCAGAATC CTCTCTTTCT 001680 001681 TCTGCCAGGG TTCTTCCTCT CCAAGCCCTC CTCTGAACCA GCTATCCCTC TGGAATGCAG CTGACCAAAC AAATTCCCAC 001760 001761 TAATGTAACT GGTAGTATCT TAGAGGTGAA CTCTCCTGTG GGCATTTTAT TTGTATCTTA GAAGAATTTT TGTTTTAAAT 001840 001841 CTTATTTTTC CCAACTGGCA TATAGTAGGG TGCTCAATAA AGGTGGGTTG CCTGAATAAG TGAAAGAACC TAAGGAACAA 001920 001921 TGTCTAATGG TGGAATTTCA GTCATGTCTT CATGCTTTCA GCCTCTCTGC AAACATTTAT TTTGTGTGCT AGATACTGGA 002000 002001 GATATGTTGC TGGATAAAAC AGCCACAGTC CTTGCCCTTT GGAACTTCCA GTATAGTGGG GACAATCAGT TAATAAGCAA 002080 002081 ACAAACACAT TTCTATTTAT AAATGGTGAT AAAATTCCTT GGCTAAAAGA ATATTGCAGA GTCTATGATA CTGTATAACA 002160 002161 GGGGCTTCTA ATTAAATTTA GGGGATCCTG ATGCCACTTA GCAGTGGGAT TTCAGAGGAC CTTTCAGAGG AGCTAAGCAT 002240 002241 TTATCAGCTG CCGCTTATTC ATTGCCTTGA TTTTCTTCAA AAGACAACTA TAATCATTCA AAAATTCCAA GTTGGGTCCA 002320 002321 AAATGTCAAA GGACTCTGCA TGCTGCTGAA AGCAAATGAA TGTGGAGGGT TTCAAACAAG ACAAAAGGCA TTCCTCCCTG 002400 002401 CCCAATCCCA GTGCAGTCAA GGCCAACCTA GGCAGCAGCC ACCTTCTCAG GAAACAAGGC CTTGGAGAAA TCAGTGCCTG 002480 002481 TGTGAAGATG GTGTCGGCTG AAAAGAAGAG CAATGGCGTC AAGTACTAAA TGATTATCAA GGGTCAACTT CTGTATCTGT 002560 002561 ATCAGCTGCA ACACACTGTC TTTAATACCC CCAGCCCAGG AGCTCGTGCC TTACATGCTG GGCCCACATT CTGCCCCAAG 002640 002641 CAGAAGCTGC AGGGACCTCA GTGTCCACCC TTCCTGCCTC ATGTCTCAAG GCAGCAGCTC CTCTCTGGTA ACCTGATGGT 002720 002721 GATGTGGCCA GGGTGCCGAG ATGGGACAGC AAAGATGATG GGCTCTGCAC ACCAACTCCA GGAAACTCAT TCTGGGCCAG 002800 002801 CTCTATGCAA AGGGAAAAGC ATGTTGCATT CACCTAGGCC AAGAAACAGT GTTGTAGGTT AAGGGCATGG ACTGCTTGAG 002880 002881 TTCAGTGCCA GCTATGCCAC TTACCAGCTG TGGGACCTTG GGCAAGTTGC CTTTGTGCCT ATGCTTCCAC TTTCCTTTTC 002960 002961 CCTCCACCAT GAAGATGGGT TAAGAGTCTC AAGGTTGTGG TGACTGTAGA TATATGAGTG TGCATTTGCT ATGCTTTACC 003040 003041 TGTCTGGCAT CTAGCAAACA TTCTTCACAT GTTTGCTGTT ATCAAGATTG TTGTATATGT GAATGAAATG ATACGATGTC 003120 003121 TCAGATTTGC TTTAAAATAA TAGGGAGGTT GGGGAAGTGG GTTGGGGTAC AGACGAAACA GGAGTAGGTG TGAGTTGATA 003200 003201 ATGTTGAAGT TGAGTGATCA GTACGTGGGT TTATTACACT ATTCTTTTCA CATTTGCTTT GAAACTGTTC ATAAAAATTT 003280 003281 CCATAATAAA AGTTCTATAT ATATATAAAT ATATACATAC ATATGAGAAA ATGATGTTTA TAGCAGAGCT TCACTAGACA 003360 003361 TGGTATGTAG CATGTAGTGC ACAAGCTGAT TTTAAGTGAT ATTCAGATGG CCTTTTAAAT TTAATTATTA TGTATTTAAA 003440 003441 ATTTAATTAA TTATAGCAAG TGATATGAAC TGTGATAATG CAGAAGGTTT CCTTTTAAAA TAAAGTTCAG TCAAAAAA |
Predicted Small Protein
| Name | ENST00000418282.1_smProtein_1502:1762 |
| Length | 87 |
| Molecular weight | 9407.8383 |
| Aromaticity | 0.116279069767 |
| Instability index | 59.4220930233 |
| Isoelectric point | 7.68414306641 |
| Runs | 11 |
| Runs residual | 0.0117146824019 |
| Runs probability | 0.0541985836104 |
| Amino acid sequence | MLGNGEVDLPCVVPPFVCPCPASWHLLFTPRAPRFSPLPRATFGLVFLFHVPACRILSFF CQGSSSPSPPLNQLSLWNAADQTNSH |
| Secondary structure | LLLLLEELLLLLLLLLLLLLLLLEEEEELLLLLLLLLLLLLLLLEEEEEELLLEEEEEEE ELLLLLLLLLLLLLLHHHHHHHLLLL |
| PRMN | LLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLLHHHHHHHHHHHHHHHHHHLL LLLLLLLLLLLLLLLLLLLLLLLLLL |
| PiMo | iiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiTTTTTTTTTTTTTTTTTToo oooooooooooooooooooooooooo |