NONHSAT081499
MIR155HG, a 1500 bp long non-coding RNA transcribed from BIC(B-cell integration cluster).
Contents
Annotated Information
APPROVED SYMBOL
MIR155HG
SYNONYMS
"B-cell receptor inducible", BIC, "BIC transcript", NCRNA00172, "non-protein coding RNA 172"
Characteristics
MIR155HG, encoding a 1500 bp non-coding RNA, is comprised of three exons and locates in cytoplasm.[1]
Cellular Localization
- Cytoplasm
Function
MIR155HG, as a T cell early activation proto-oncogene, might act to alter the genetic program, such that upon antigen re-stimulation.[2]
Regulation
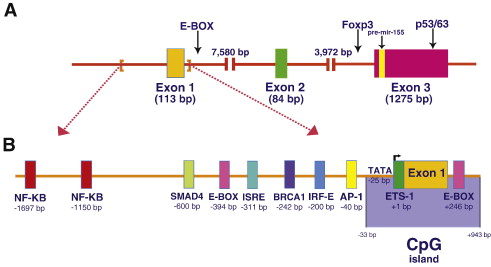
MIR155HG promoter may be activated by both AP-1- and NF-κB-mediated mechanisms upon B-cell stimulation[1].
Diseases
- B-cell chronic lymphocytic leukemia, Breast cancer [3]
Expression
MIR155HG expression is highly induced by antigen receptor stimulation of B-cells (i.e. B-cell receptor cross-linking) [3]
Labs working on this lncRNA
- Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH, USA.[1]
- First Faculty of Medicine and Center of Experimental Hematology, Charles University in Prague, Prague, Czech Republic.[3]
- Global Pharmaceutical Research and Development, Abbott Laboratories, Abbott Park, IL 60064, USA.[2]
References
<references> [3](1) [1](2) [2](3)
Basic Information
| Transcript ID |
NONHSAT081499 |
| Source |
NONCODE4.0 |
| Same with |
MIR155HG, BIC |
| Classification |
intergenic |
| Length |
5409 nt |
| Genomic location |
chr21+:26934386..26951344 |
| Exon number |
3 |
| Exons |
26934386..26934569,26942149..26942233,26946205..26951344 |
| Genome context |
|
| Sequence |
000001 CTGGTCGGTT ATGAGTCACA AGTGAGTTAT AAAAGGGTCG CACGTTCGCA GGCGCGGGCT TCCTGTGCGC GGCCGAGCCC 000080
000081 GGGCCCAGCG CCGCCTGCAG CCTCGGGAAG GGAGCGGATA GCGGAGCCCC GAGCCGCCCG CAGAGCAAGC GCGGGGAACC 000160 000161 AAGGAGACGC TCCTGGCACT GCAGATAACT TGTCTGCATT TCAAGAACAA CCTACCAGAG ACCTTACCTG TCACCTTGGC 000240 000241 TCTCCCACCC AATGGAGATG GCTCTAATGG TGGCACAAAC CAGGAAGGGG AAATCTGTGG TTTAAATTCT TTATGCCTCA 000320 000321 TCCTCTGAGT GCTGAAGGCT TGCTGTAGGC TGTATGCTGT TAATGCTAAT CGTGATAGGG GTTTTTGCCT CCAACTGACT 000400 000401 CCTACATATT AGCATTAACA GTGTATGATG CCTGTTACTA GCATTCACAT GGAACAAATT GCTGCCGTGG GAGGATGACA 000480 000481 AAGAAGCATG AGTCACCCTG CTGGATAAAC TTAGACTTCA GGCTTTATCA TTTTTCAATC TGTTAATCAT AATCTGGTCA 000560 000561 CTGGGATGTT CAACCTTAAA CTAAGTTTTG AAAGTAAGGT TATTTAAAAG ATTTATCAGT AGTATCCTAA ATGCAAACAT 000640 000641 TTTCATTTAA ATGTCAAGCC CATGTTTGTT TTTATCATTA ACAGAAAATA TATTCATGTC ATTCTTAATT GCAGGTTTTG 000720 000721 GCTTGTTCAT TATAATGTTC ATAAACACCT TTGATTCAAC TGTTAGAAAT GTGGGCTAAA CACAAATTTC TATAATATTT 000800 000801 TTGTAGTTAA AAATTAGAAG GACTACTAAC CTCCAGTTAT ATCATGGATT GTCTGGCAAC GTTTTTTAAA AGATTTAGAA 000880 000881 ACTGGTACTT TCCCCCAGGT AACGATTTTC TGTTCAGGCA ACTTCAGTTT AAAATTAATA CTTTTATTTG ACTCTTAAAG 000960 000961 GGAAACTGAA AGGCTATGAA GCTGAATTTT TTTAATGAAA TATTTTTAAC AGTTAGCAGG GTAAATAACA TCTGACAGCT 001040 001041 AATGAGATAT TTTTTCCATA CAAGATAAAA AGATTTAATC AAAAAATTTC ATATTTGAAA TGAAGTCCCA AATCTAGGTT 001120 001121 CAAGTTCAAT AGCTTAGCCA CATAATACGG TTGTGCGAGC AGAGAATCTA CCTTTCCACT TCTAAGCCTG TTTCTTCCTC 001200 001201 CATATGGGGA TAATACTTTA CAAGGTTGTT GTGAGGCTTA GATGAGATAG AGAATTATTC CATAAGATAA TCAAGTGCTA 001280 001281 CATTAATGTT ATAGTTAGAT TAATCCAAGA ACTAGTCACC CTACTTTATT AGAGAAGAGA AAAGCTAATG ATTTGATTTG 001360 001361 CAGAATATTT AAGGTTTGGA TTTCTATGCA GTTTTTCTAA ATAACCATCA CTTACAAATA TGTAACCAAA CGTAATTGTT 001440 001441 AGTATATTTA ATGTAAACTT GTTTTAACAA CTCTTCTCAA CATTTTGTCC AGGTTATTCA CTGTAACCAA ATAAATCTCA 001520 001521 TGAGTCTTTA GTTGATTTAA AATAATTTTT ATGGCCCATT TTCTTTTGCA AAGTACAAAT TATAATTCCA GAAGCTATCT 001600 001601 GCCAAAAAAA GACAATGGAA GAAAATTATG AGGACACTAA AGGGAACATT CCTGACTCCT AGGACACAGT ATTCTCATGT 001680 001681 AAGAATACCC ATTTGGCTTT GCCCCACTTT CTGTTCAAGT TGTATGTGCT GCCAAGACAA ATTTCTTCTC CTCACTTTCT 001760 001761 GTTCAAGTTG TATGTGCTGC CAACACAAAT TTCTTCTCCT AACACCTGGA ATGAGCTTGT GGCTCCACGG AGCTCCCATG 001840 001841 TAGGTAGAAC ACATTCAAGA GTGTTTCTTC AGGCCAGGCA CAGTGGCTCA CGCCTATAAT TCCAACACTT TGGGAGGCCA 001920 001921 AGGCGGGTGG TTCACTTGAG GTCAGGAGTT CTACACTACC CTGGCCAACA TAGTGAAACC TCGTCTCTAC TAAAAATACA 002000 002001 AAAATTAGCT AGGCATGGTG GTGCGTGCCT GTAATCCCAG ATACTCAGGA GGCTGGGGCA GAAGAATCAC TTGAACCCAG 002080 002081 GAGGTGGAGT TCACAGTGAA CGAAGATCGT GCCACTGCCC TCCAGCCTGG GTGACAGAGT GAGACTCCAT CTCAAAAAAA 002160 002161 AAAAAAAGTG TCTGTTCAAT TCATTTTATC TGCCACTGTA TATCTACAAG AGGACTCCTT GCTTTGAGTC CCAGTTCTTC 002240 002241 AAAATCCTGC TTGCTGCCTA AAGACACAAA GTCATAGAAC ATTAGGACTG GAATAAACTA TTGTAGTATA ACCTTCCTGC 002320 002321 TGTTACAATA AAAAGCTATA AACATTACAA AGGCACTTAA AGAAAAGCTG TGCTGGGAAA TGCCTTATTT GAAGAGCATT 002400 002401 TGGGTACACT CACATGAAGC TTTGGAAAGT TACTTAATCT TCCTCTAAGC CTCAATTTCT TCATCAATAA AATGATGAAT 002480 002481 ATATTTCATA TTTCATAAGA TGTAGGCTTA AATGAAATCA TGCATGTGAA AAGCACAATG TCTATTATGC ATTAAGTGTT 002560 002561 AAACACAAAC TTTCAAACTT TTACATTTCA GTCATTTAAA TCAAATGATC TACCATAGCA GATCTCCCCA AAGACACTCT 002640 002641 AGCTCCAAAC TAACTATAAT CAATCCCGTG GTGACGACCC AAAGAACCAG GGTTCCTGGG TTTTCTCTCA CGCCCAGCCC 002720 002721 ACCTAGGCTT CTGACTGCCT ATCTCCTCAT CTTAGAGGGA TTCCTCCTGA CCAGTCATTA CAAAAATTCT TGGAGCATGA 002800 002801 GCAAGACTGA GTCTGTGTTT GGATTGGAGG AAGGGGTAAG TAACAAGGTA TATATATGTA TATGTGTGTG TGTGTGTGTG 002880 002881 TGTGTGTGTG TGTGTGTGTG CGCGCGCGTT TGTGTTTAAG TTACAAACTA AGGATGGTTT TAAAAAATTG CAAAATGTTA 002960 002961 AGAGATTTAT CTAGGATCTG ACCCACTTCC CCAGAAATTT ACGGAATTCA AGACCAAGCC TTTCGTATTC ACTGTCATCA 003040 003041 GTTAGATGGA CCCAGTAGCT AAAAAATTAA GGAGAAAAAA AATAGAGGAA TATGAGGGAA AAAGTCAAGT CAGAGGACTG 003120 003121 AAGAATATAT ACAGAACATA GAATATTAGA AATTTTCTGA AAATAGATGA TAGCAAATAC CTCAAGAAAG TGAGGAAACG 003200 003201 TGAAAAGGGA GCTTAAAGAA AAATAATAGA GTCCATACAT GGGACACAAG CACAAAAAGC TGACTGAGCA TGTGGTTCTC 003280 003281 TACTAATAAT AACTGGAAAA CAGTGAAACC AGCAGGAAAA ACTTGCACAT AAGAGTTTAA CTTTCCACTT TGGGTTTCAA 003360 003361 ATTCTCCCAT AGCAATAAGA GTAAGTGAAA TAATGTTCAC ATAAGGATTT TTTTTTTTTC TATTACTGTA GTGTTTTGTT 003440 003441 TTGTTTTGTT TTGGTTTTGG TTTTTGGTGG TTTTTTTTTT TTTGAGACAA AGTCTCGCTC TTGTCCCCCA GTCTGGAGTG 003520 003521 CAATGTCATG ATCTTGGCTC ACTGCAACCT CCGCCTCCCG GGTTCAAGTG ATTCTCCTGC CTCAGCCTCC CAAGTGGCTG 003600 003601 GGATTACAGG TGCGTACCAC CACGCCCAGC TAATTTTTTG TATTTTAAGC AGAGACGGGG TTTCACCATG TTGGCCAGGC 003680 003681 TGGTCTCGAA CTCCTGACCT CAGGTGATCC ATCCATCTCA GCCTCCCAAA GTGCTGGGAT TACAGGTGTG AGCCACCGCA 003760 003761 CCCGGCCTGT TACTGTAGTG TTTTAACAAA TCTAAGTGTA ATAGTATTAG TCTTCATTCA TCTAAGACAT ACAAGTACTG 003840 003841 GAATTTTTTA GGTGGGTTCC TCAAATGTCA TCACAGTGGT GTCTTCTCTA CATTCAGTCA CTCTTCCGCC CTTCTCTGTA 003920 003921 ATGCTCAGGG ATTTCCCAGT TCACTAGCAC TTAACATAGT ATGTCCTTAC TTACTTTGGG GTATAAATGA CTGCAATAGA 004000 004001 AGAAGTGATA TCCTTGAGGG ATGAAACACG ATCTTGCATA TTCATATCCC TTTCAAATGT TTACTGACTT AATCACACCA 004080 004081 TTAGTTTTCT AAGTCCAAGT CTAAAAACAG AAATGAGTTT AAATACAGTG AACATTTCAT TTTCACTATA CCACTAAGCT 004160 004161 GAAAGAAATG ATTTTTGGTA AATAAAAGTT CATCACAGTG AAAAGTTCTA CATATAACTG AAGATCAAAG GATTCTCACC 004240 004241 TGGGAAGCTT GTCAAGATGC ATGTTCCCAA GCTCCCAGGC TCCCACTCCC AGAGATTCTA ATACTGTAAT GGAGCTGAGG 004320 004321 ACACTACATT TTTAACAGGC TGACCAGGTA GTTCAGATTC AGGTGGTTCC TAAACCAACT CTGAGGAAGT AGTGGTTTTA 004400 004401 TATACGATGC TACTGTTGAT GTGGCTACTA GGAATGGTGC TCAGCAGAAA TCCTGCAACA TATGGATCTG TTTACACACT 004480 004481 ATTCACTGTG TAATGAGAGT CTGCAACATA GGCTAAGTAA TAAGAGATGT CTTGTGGTTG ACCTCTCTGG AGATAAGCAG 004560 004561 CAATGAAATT CCAATGAAGT TTCATCATCT CTCATCACCA AAACCATCCT AACCACCTCA ATAAAATACA CCTGACTTCC 004640 004641 CCCGCTCCCC CGACCCACCA CCCCAAAACC AGCTGCCCTA TGTTCTACCA AAAGCTGCAA CCAACCAATG ACCTGATACT 004720 004721 AACCCCAATC TATCAGCTGC TGCCTAGTGC AACAGAGAGG GAAAGTCAAT GAGAAGGAAT TGGGTGCCAG TTCTTGCTTT 004800 004801 GCAAGGAGTG GGTTGACTAA TGAGACAGCC ACAAATGTCA CTGCCATCCC ATCTCACCCT TTCACCTGTG CTGCAAGGCT 004880 004881 TCAACAATCA TACCCAGTCT CCACTTAGGA AAATTCTGTC CTTTCTCAAA TGCTGGCCAC TCAGCAATGT GCTCCCTTAA 004960 004961 TCCAGGCCAA GCACATAGTA AAAGAACTCC AAAAAGGGTT ACAGCCAAAA GTGATCTTTC TCCTCCATAC TCCCAAAGCA 005040 005041 GATTATAGCT CCTCCATAAA CGTGCTTCGC ATTGTGCCCC ACGTTTGGTT TGTTACTTGT GTATACATTA TCGTCTCTTC 005120 005121 TAGATTGAGC TTCTGTAGGA AAGGAGAATC CTTTTCAATT TTTTGGTATT ACACCCAGCT CAGGACCTTG CTCAGTTGCT 005200 005201 TAAATTGAAT GAGAAAATGT ATCTAAGTAC CCACTGATGT TGGAAATTCA ACTTGTTCCA ATATTGAGGA CTGCCTATAC 005280 005281 CTGGCATATG CGTGTCCCAA TTTGAGAAAT TGTTTTTCAA GTTGACTTGT ATGGATTCCT TACAGACGCT CAAGCATCAG 005360 005361 TTTGGTTTTC CCTAAATCCT GCAGATCAGG AAGGCAGTAT CCATGTTTG |
- ↑ 1.0 1.1 1.2 1.3 1.4 Elton, T. S., H. Selemon, S. M. Elton, and N. L. Parinandi. "Regulation of the Mir155 Host Gene in Physiological and Pathological Processes." Gene 532, no. 1 (2013): 1-12.
- ↑ 2.0 2.1 2.2 Haasch, D., Y. W. Chen, R. M. Reilly, X. G. Chiou, S. Koterski, M. L. Smith, P. Kroeger, K. McWeeny, D. N. Halbert, K. W. Mollison, S. W. Djuric, and J. M. Trevillyan. "T Cell Activation Induces a Noncoding Rna Transcript Sensitive to Inhibition by Immunosuppressant Drugs and Encoded by the Proto-Oncogene, Bic." Cell Immunol 217, no. 1-2 (2002): 78-86.
- ↑ 3.0 3.1 3.2 3.3 Vargova, K., N. Curik, P. Burda, P. Basova, V. Kulvait, V. Pospisil, F. Savvulidi, J. Kokavec, E. Necas, A. Berkova, P. Obrtlikova, J. Karban, M. Mraz, S. Pospisilova, J. Mayer, M. Trneny, J. Zavadil, and T. Stopka. "Myb Transcriptionally Regulates the Mir-155 Host Gene in Chronic Lymphocytic Leukemia." Blood 117, no. 14 (2011): 3816-25.